Lab 5: Contrasts, Estimation, and Power Analysis
FANR 6750: Experimental Methods in Forestry and Natural Resources Research
Fall 2022
lab05_contrasts.RmdContrasts
Mussel data
library(FANR6750)
data("musselData")
head(musselData)
#> Watershed Length
#> 1 Twelvemile 16
#> 2 Twelvemile 8
#> 3 Twelvemile 12
#> 4 Twelvemile 17
#> 5 Twelvemile 13
#> 6 Chattooga 18
# By default, R will order watershed in alphabetical order
# We will change it to a factor and explicitly set the order to match the order from lecture
musselData$Watershed <- factor(musselData$Watershed,
levels = c("Twelvemile", "Chattooga","Keowee", "Coneross"))| Watershed | Length |
|---|---|
| Twelvemile | 16 |
| Twelvemile | 8 |
| Twelvemile | 12 |
| Twelvemile | 17 |
| Twelvemile | 13 |
| Chattooga | 18 |
Questions
Suppose we want to make 3 a priori comparisons:
Agricultural (Twelvemile & Coneross) vs. Forested (Chattooga & Keowee)
Within agricultural (Twelvemile vs Coneross)
Within forested (Chattooga vs Keowee)
| Comparison | Null hypothesis | |
|---|---|---|
| 1 | Ag. vs Forested | \(\frac{\mu_T + \mu_{Co}}{2} - \frac{\mu_{Ch} + \mu_{K}}{2} = 0\) |
| 2 | Twelvemile vs Coneross | \(\mu_T - \mu_{Co} = 0\) |
| 3 | Chattooga vs Keowee | \(\mu_{Ch} - \mu_{K} = 0\) |
Constructing contrasts in R
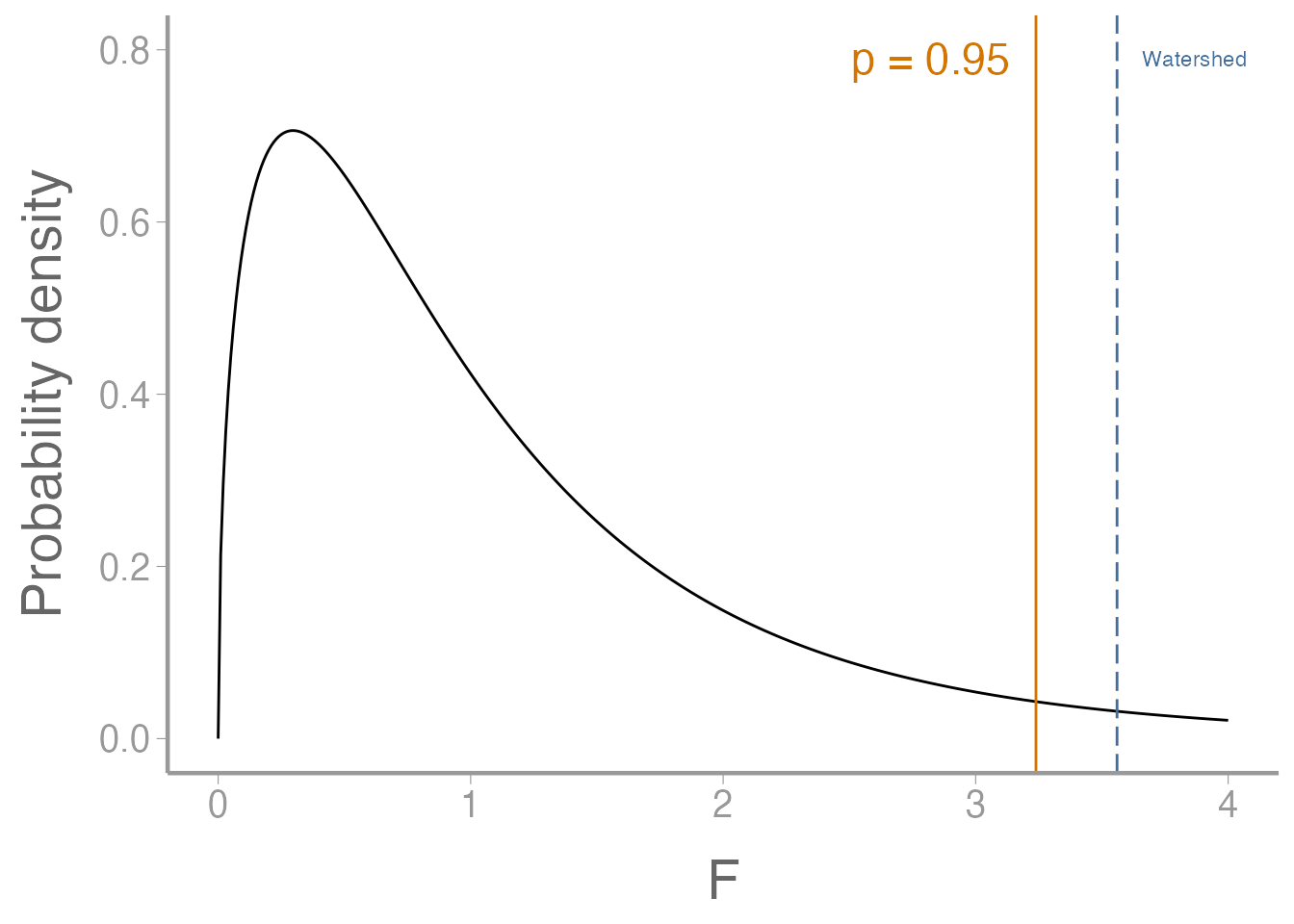
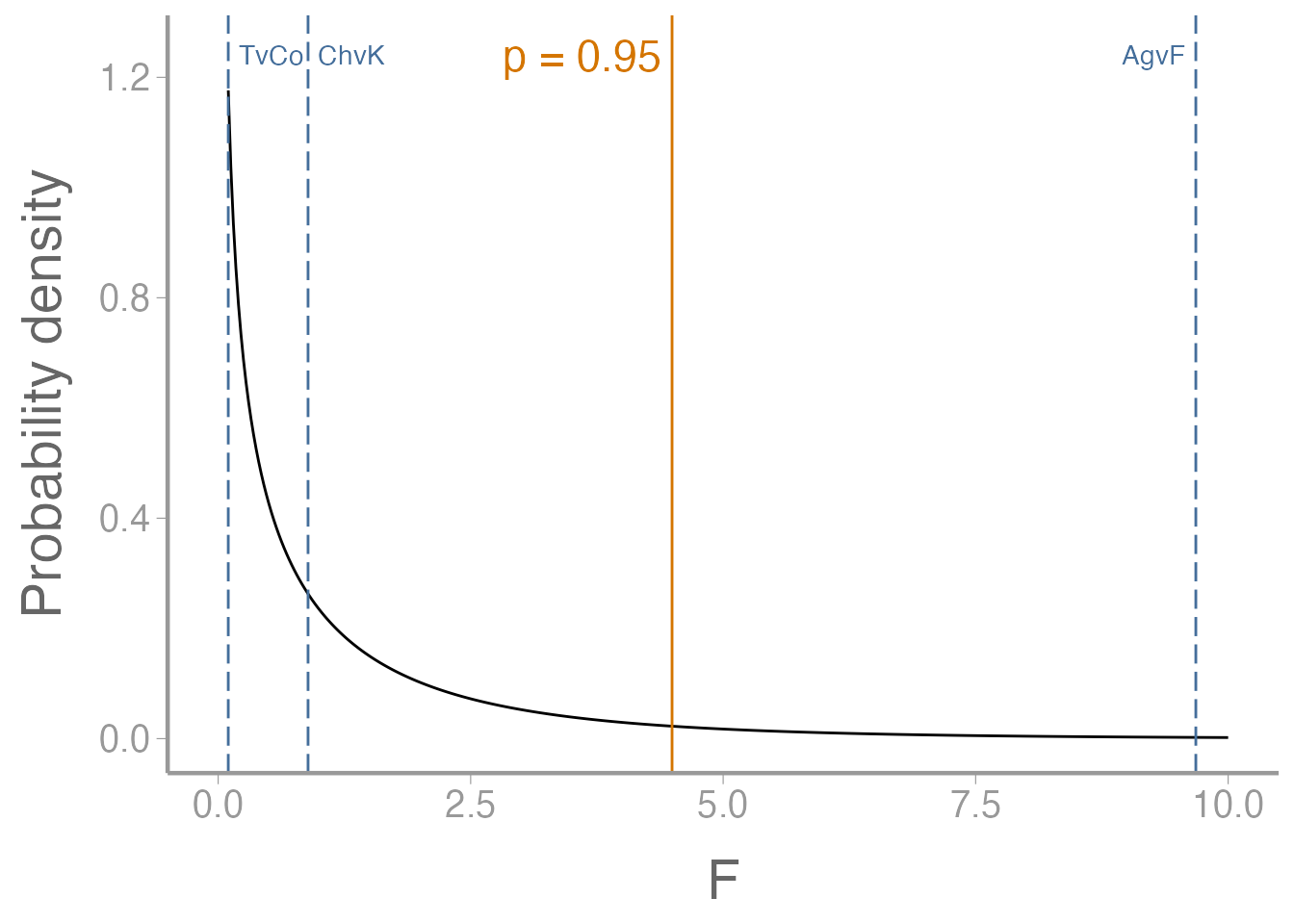
Are they orthogonal?
Step 1: sum of coefficients is 0, for each contrast.
all() tests whether all its arguments evaluate to
TRUE
Step 2: pairwise dot products are all equal to 0.
Step 2 (alternate method): pairwise dot products are all equal to 0.
Try the %*% operator:
AgvF %*% TvCo
#> [,1]
#> [1,] 0When the operands are vectors, %*% does the dot
product
Contrasts
Now partition the sum-of-squares with split=
argument:
summary(aov.out,
split = list(Watershed = c("AgvF" = 1,
"TvCo" = 2,
"ChvK" = 3)))
#> Df Sum Sq Mean Sq F value Pr(>F)
#> Watershed 3 393 131 9.70 0.00069 ***
#> Watershed: AgvF 1 361 361 26.76 9.2e-05 ***
#> Watershed: TvCo 1 12 12 0.90 0.35786
#> Watershed: ChvK 1 20 20 1.45 0.24575
#> Residuals 16 216 14
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Standard errors for each contrast
SE for Twelvemile vs Coneross
se.contrast(aov.out,
contrast.obj = list(Watershed == "Twelvemile", Watershed == "Coneross"),
data = musselData)
#> [1] 2.324SE for Chattooga vs Keowee
se.contrast(aov.out,
contrast.obj = list(Watershed == "Chattooga", Watershed == "Keowee"),
data = musselData)
#> [1] 2.324SE for Ag. vs Forested
se.contrast(aov.out,
contrast.obj = list(Watershed %in% c("Twelvemile","Coneross"),
Watershed %in% c("Chattooga","Keowee")),
data = musselData)
#> [1] 1.643The %in% function returns TRUE for any
elements of the first vector that match an element in the second vector
and FALSE otherwise
Estimation
Estimating confidence intervals
In an ANOVA context, confidence intervals can be constructed using the equation:
\[CI = Point\; estimate \pm t_{\alpha/2,a(n−1)}\times SE\] As usual, the hard part is computing the SE
Confidence intervals from one-way ANOVA
SE’s for the effect sizes (\(\alpha\)’s)
(effects.SE <- model.tables(aov.out, type="effects", se=TRUE))
#> Tables of effects
#>
#> Watershed
#> Watershed
#> Twelvemile Chattooga Keowee Coneross
#> -5.35 2.85 5.65 -3.15
#>
#> Standard errors of effects
#> Watershed
#> 1.643
#> replic. 5Extract the \(\alpha\)’s and the SEs
# str(effects.SE)
alpha.i <- as.numeric(effects.SE$tables$Watershed)
SE <- as.numeric(effects.SE$se)Compute confidence intervals
tc <- qt(0.975, 4 * (5 - 1))
lowerCI <- alpha.i - tc * SE
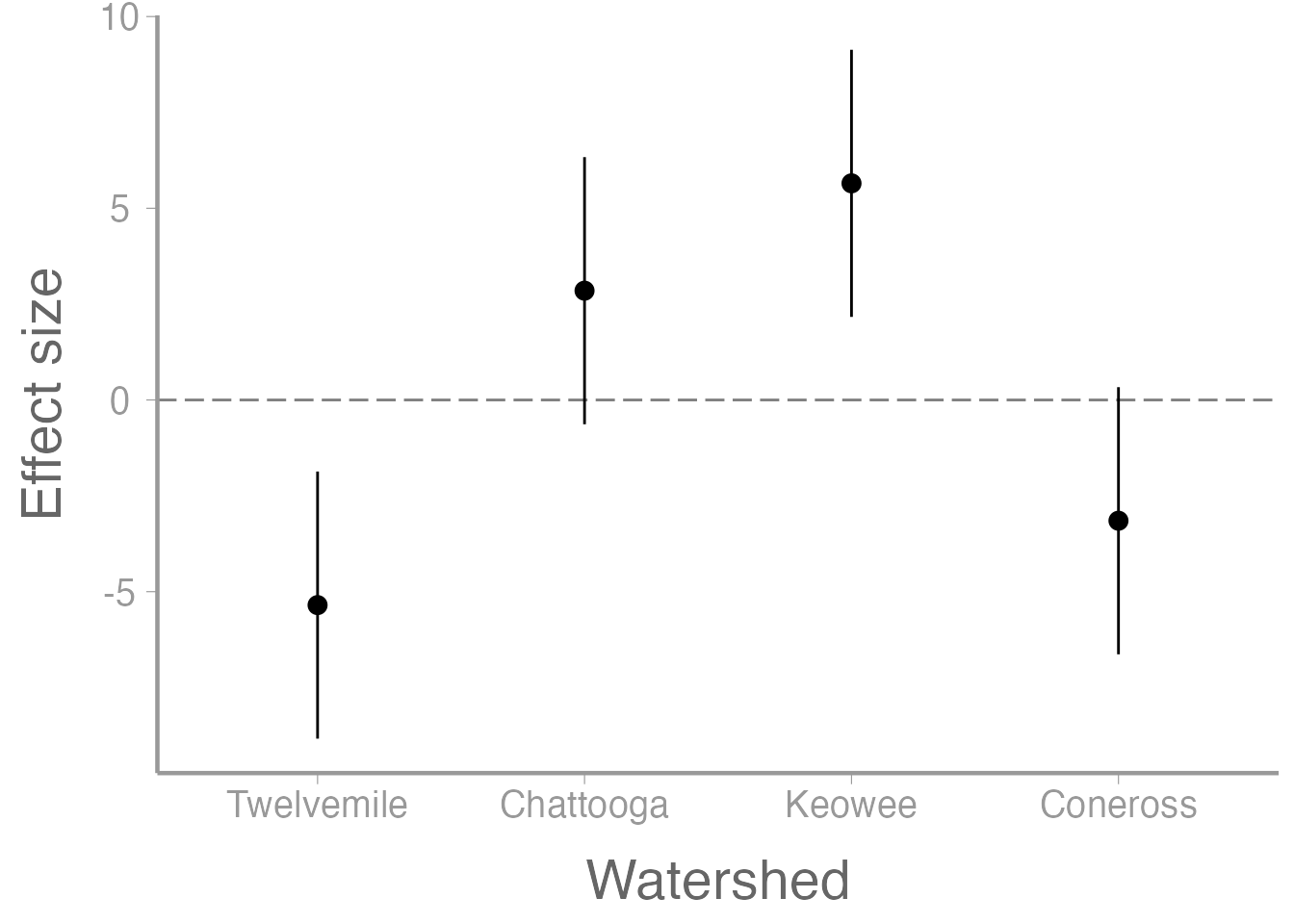
upperCI <- alpha.i + tc * SEPlot effects and CIs
Put results into a data frame
CI <- data.frame(Watershed = factor(c("Twelvemile", "Chattooga", "Keowee", "Coneross"),
levels = c("Twelvemile", "Chattooga","Keowee", "Coneross")),
effect.size = alpha.i,
SE,
lowerCI,
upperCI)
head(CI)
#> Watershed effect.size SE lowerCI upperCI
#> 1 Twelvemile -5.35 1.643 -8.8334 -1.8666
#> 2 Chattooga 2.85 1.643 -0.6334 6.3334
#> 3 Keowee 5.65 1.643 2.1666 9.1334
#> 4 Coneross -3.15 1.643 -6.6334 0.3334Power analysis
Statistical Power
Errors:
Type I: the null hypothesis is true, but you reject it
Type II: the null hypothesis is false, but you fail to reject it
\(\alpha = Pr(Type\;I\;error)\)
\(\beta = Pr(Type\;II\;error)\)
Power = Pr(rejecting a false null) = \(1 − \beta\)
In R functions power.t.test() and
power.anova.test(), you provide all but one of the
“ingredients” for the hypothesis test and determination of power. Then
the missing ingredient is estimated.
Power analysis for a 2-sample t-test
power.t.test(n = NULL,
delta = 3,
sd = 2,
sig.level = 0.05,
power = 0.8)
#>
#> Two-sample t test power calculation
#>
#> n = 8.06
#> delta = 3
#> sd = 2
#> sig.level = 0.05
#> power = 0.8
#> alternative = two.sided
#>
#> NOTE: n is number in *each* groupPower analysis for one-way ANOVA
power.anova.test(groups = 4,
n = 5,
between.var = 360.0,
within.var = 101.2,
power = NULL)
#>
#> Balanced one-way analysis of variance power calculation
#>
#> groups = 4
#> n = 5
#> between.var = 360
#> within.var = 101.2
#> sig.level = 0.05
#> power = 0.9999
#>
#> NOTE: n is number in each groupAssignment (not for a grade)
Researchers wish to know if food supplementation affects the growth of nestling Canada warblers. The treatment groups are: (A) No supplementation control, (B) low, (C) medium, (D) high, and (E) very high. The response variable is the weight of a ten-day-old nestling.
Create an R Markdown file to do the following:
- Create an
Rchunk to load the data using:
-
The researchers are interested in the following contrasts. Create a header called “Contrasts” and determine whether these contrasts are orthogonal. Show your work!
Groups A,B vs C,D,E
Groups A vs B
Groups C vs D,E
Groups D vs E
Create a header called “ANOVA analysis”. Under this header, use the
warblerWeightdata to test the null hypothesis of each contrast. This section should include a well-formatted ANOVA table showing the results of the analysis. Use thecaptionargument in thekable()function to include a brief caption for the ANOVA table and theroundargument to print an appropriate number of digits.-
In addition to the ANOVA table, compute and report, in a second table (with caption), the following for each contrast:
The difference in the means
The SE of the difference in means
The 95% CI for the difference in means
Suppose you wanted to replicate the study with a smaller sample size of \(n = 2\) per treatment group. In a new section called “Power analysis”, calculate and report your power for this new experiment. This section should include both the
Rcode you used to conduct the power analysis, and a brief (1-3 sentence) description of the result and interpretation.
A few things to remember when creating the document:
Be sure the output type is set to:
output: html_documentBe sure to set
echo = TRUEin allRchunks so that all code and output is visible in the knitted documentRegularly knit the document as you work to check for errors
See the R Markdown reference sheet for help with creating
Rchunks, equations, tables, etc.See the “Creating publication-quality graphics” reference sheet for tips on formatting figures